You can follow the steps described in this tutorial to become familiar with the procedure for data reduction of EPMA data and to discover the main features of XMapTools. A video tutorial is available on Youtube:
Located in China? Watch this video on Bilibili
Table of Contents
Dataset description
The dataset used in this tutorial comes from the study of Duesterhoeft & Lanari (2020). It consists of EPMA maps of the sample HAL104 from West Guilford (Ontario) of the Canadian Grenville Province. It was analysed using 15 KeV accelerating voltage, 100 nA specimen current, 130 ms dwell time, 20 µm step size, ~1 µm beam size with 1000*1000 pixels. The mapping time was ~75 h.
Please do not use these data for any other purpose than this tutorial without the explicit authorisation of the author(s).
| File | Type | Description |
|---|---|---|
| _Ce.txt | EDS map | Mostly bellow detection limit |
| _Cr.txt | EDS map | Mostly bellow detection limit |
| _La.txt | EDS map | Mostly bellow detection limit |
| _Ni.txt | EDS map | Bellow detection limit |
| _P.txt | EDS map | Overlap with Zr |
| _S.txt | EDS map | |
| Al.txt | WDS map | |
| Ca.txt | WDS map | |
| Fe.txt | WDS map | |
| K.txt | WDS map | |
| Mg.txt | WDS map | |
| Mn.txt | WDS map | |
| Na.txt | WDS map | |
| SEI.txt | WDS map | Secondary electron image |
| Si.txt | WDS map | |
| Ti.txt | WDS map | |
| TOPO.txt | WDS map | Topographic image (obtained by BSE) |
| Standards.txt | Standard file | File containing the spot analyses used for map standardisation. |
Reference: Duesterhoeft, E. & Lanari, P. (2020). Iterative thermodynamic modelling – Part 1: A theoretical scoring technique and a computer program (BINGO-ANTIDOTE). Journal of Metamorphic Geology, 38, 527-55.
Tutorial
How to get started?
- Download Tutorial_EPMA_2023.zip from the download center
- Unzip the folder in a suitable working directory (e.g. Documents/Mapping/EPMA/Tutorial_EPMA_2023/)
- Open XMapTools. Note that starting XMapTools can take up to a minute; restarting the program is slightly faster
- When the program is ready, a dialog box opens; pick the working directory Tutorial_EPMA_2023 and click the button “Open”. You can change the working directory later, but you need to pick a directory when you start the program. A log file is automatically created in the selected working directory.
Import maps
- 0In the first workspace “PROJECT & IMPORT”, click on the button “Import”
 (Import Maps) to open the Import Tool module
(Import Maps) to open the Import Tool module - Select all the map files to be imported and click on the button “Open”. Note that you can select all the text files available in the folder and XMapTools will find out which ones can be imported as maps
- In the tab “Corrections”, change the value of dwell time to 120 ms
- Press the button “Import Data”
X-ray data visualization (Part 1)
- Quickly go through the maps and use the auto-contrast option
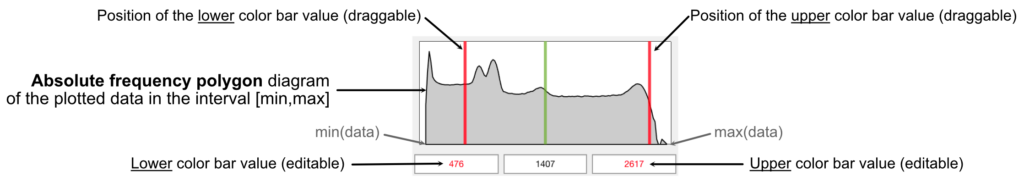
 or the live frequency polygon (see figure bellow) to adjust the lower and upper limits of the color bar. Note that the color bar settings are available in the workspace “Options”.
or the live frequency polygon (see figure bellow) to adjust the lower and upper limits of the color bar. Note that the color bar settings are available in the workspace “Options”. - Eliminate the maps which are not needed because they do not contain any important information (Ce, Cr, Ni, S). To eliminate a map, select the map in the primary tree menu, then right click on the name and select “Delete”
- Save your project using either the button “Save” available in the workspace “PROJECT & IMPORT” or in the menu “File” > “Save Project”
Classification
Classification is the step used to create a classified image in which each pixels are attributed to a class (mineral, glass, epoxy, etc.). In this tutorial we are using the random forest algorithm. The classification in XMapTools is supervised. You need to create a “training set” containing training data for each class.
- Set the active workspace to “CLASSIFY”
- Create a new training set. In the secondary tree menu, select the category “Training Set (Classification)” and press the button “Add”
 available in the workspace “CLASSIFY” to add phase definitions. Select in the list the phases: Biotite, Garnet, Plagioclase, Quartz; then click “Apply”
available in the workspace “CLASSIFY” to add phase definitions. Select in the list the phases: Biotite, Garnet, Plagioclase, Quartz; then click “Apply” - Add training data for each phase:
- Select a training class in the training set
- Adjust display (optional): select an element to be display and eventually use the zoom
 and pan
and pan  features to adjust the field of view
features to adjust the field of view - Select a ROI type in the dropdown menu among “Rectangle ROI”, “Polygon ROI”, “Ellipse ROI” and “Circle ROI”
- Finally, press the button “Add a ROI”
 and immediately click and drag (or click for polygon) over the map to create the ROI. How to draw a ROI in XMapTools:
and immediately click and drag (or click for polygon) over the map to create the ROI. How to draw a ROI in XMapTools:
- Rectangle: click to select the first corner and drag the mouse to the opposite corner defining a rectangle
- Polygon: click successively on the image to draw a polygon; right-click or double-click to validate and close automatically the shape
- Ellipse: click to select the first point and drag the mouse to a second point defining the long axis of an ellipse
- Circle: click to select the first corner of a square and drag the mouse to opposite corner defining a circle.
- Edit the shape of a ROI by selecting the ROI in the secondary tree menu. If the editing mode is disabled, press once the “Ctrl” key on your keyboard and try again editing the shape of the ROI.
- You can eliminate a ROI by first selecting the ROI in the secondary tree menu and then right clicking on its name and selecting “Delete”
- Add new phase definitions for sillimanite and sudoite (low temperature chlorite here replacing cordierite). Add training data for each phase (see step 3 above).
- Restore the view using either the button “Reset Zoom & Pan”
 or the button “Restore view” available near the top-right corner of the main figure
or the button “Restore view” available near the top-right corner of the main figure - In classification parameters:
- Add all maps in the list for classification. Select an element to display in the primary tree menu and use the button
 to add all maps at once
to add all maps at once - Eliminate La from the list; select La_EDS in the primary tree menu and press the button “Take La_EDS out”

- You can calculate PCA maps using the function “Generate Maps of the Principal Component”
 . The maps of principal components are listed in the category “Other” available in the primary tree menu
. The maps of principal components are listed in the category “Other” available in the primary tree menu - Select the data type “Maps” (other data type will be described in a publication)
- Select the map scaling method “Robust”; select the option “reproducibility” and set the seed value to 1
- Add all maps in the list for classification. Select an element to display in the primary tree menu and use the button
- Select the algorithm “Random Forest” and set the option corresponding to the maximum number of trees to 50
- Select the phase definition object “PhaseDef_1” in the secondary tree menu and press the button “Classify”

- Select a mask file to display the mineral map
- Check the results of the classification (for Random Forest):
- Are all training pixels classified in the true class? If not, the training set should be adjusted accordingly
- Is the out-of-bag classification error reaching a value of zero when the number of grown trees is exceeding 20? If not, there is probably a problem in the phase definition
- Check the predictor importance; maps with a very low predictor importance can be eliminated without affecting the quality of the classification (optional)
- Evaluate the quality of the classification by displaying 1-2 diagnostic elements for each class (e.g. Si for biotite, quartz, plagioclase)
- If necessary, adjust the training set and perform a new classification. It is recommended to eliminate the mask files that are not used
- Save your project
Extracting mineral modes
- Select a mask file
- Select the ROI shape “rectangle” using the dropdown menu in “Mask analysis and visualization”
- Press the button “Add a ROI to export mask modes”
 and immediately click and drag over the map to create the ROI
and immediately click and drag over the map to create the ROI - Adjust the shape to obtain the modes of the entire map. If the editing mode is disabled, press once the “Ctrl” key on your keyboard and try again editing the shape of the ROI.
- Save the modes using the button save
 in the composition tab
in the composition tab - Note that you can also copy the data to the clipboard using the button copy
 . Paste the data in a spreadsheet software program
. Paste the data in a spreadsheet software program - Eliminate the ROI using the function “Reset ROI”

- Calculate pseudo modes using the point counting method:
- Set the number of points to 500
- Unselect the Monte-Carlo option
- Extract several successive sets of pseudo modes using the point counting method by clicking on the button
 and copy the results in a spreadsheet software program
and copy the results in a spreadsheet software program
- Calculate pseudo modes using the point counting method and Monte Carlo:
- Set the number of points to 300
- Select the Monte-Carlo optionExtract pseudo modes and uncertainties using the button
 and copy the results in a spreadsheet software program
and copy the results in a spreadsheet software program - You can repeat this calculation for several number of points (e.g. 500, 750, 1000) and copy the results in a spreadsheet software program
Analytical standardisation
The analytical standardisation consists of transforming the intensity maps of elements into weight percent maps of oxides. XMapTools 4 includes a calibration module that performs an automated calibration of all minerals. This black box algorithm works ok when all phases have spot analyses to be used as internal standard and when intensities are heigh enough. However, you need to carefully check the results of the calibration for each mineral.
- Set the active workspace to “CALIBRATE”
- Display an intensity map showing a maximum of minerals (e.g. Al)
- Select the option “Import standards.txt” in “EPMA STANDARD DATA”
- Click on the button Import
 . Note that the color of the spot labels can be changed in the options of the workspace “Project & Import”
. Note that the color of the spot labels can be changed in the options of the workspace “Project & Import” - Select the element Mg to estimate the quality of the position. The diagrams on the right show correlation maps with the center of the image being the position of the spots on the map. In this case the original position is good and there is no need to change it. (Optional) you can try to shift the spot position in the correlation plot of the element and then to press “apply”
 . To restore the original position, you can simply load the standard data again using the import function
. To restore the original position, you can simply load the standard data again using the import function 
- Add two additional internal standard analyses for quartz (not measured)
- Display the silicon map by selecting Si in the primary tree menu
- Select the category “Standards (Spots)” in the secondary tree menu
- Adjust the view using zoom
 and pan
and pan  (optional)
(optional) - Press the button Add
 available in the workspace “CALIBRATE” select a pixel of quartz by clicking on the main image.
available in the workspace “CALIBRATE” select a pixel of quartz by clicking on the main image. - Set the composition to 100 wt% of SiO2 in the table available in the “Standard” tab
- Note that the new standard is listed at the bottom of the secondary tree menu under “Standards (Spots)”. Selecting this internal standard display the location and the composition.
- Calibrate the intensity element maps into maps of wt% of oxide:
- Select a mask file in the secondary tree menu
- In the workspace “CALIBRATE”, press the button “Calibrate”
 to open the Calibration assistant for EPMA data
to open the Calibration assistant for EPMA data - Check the calibration curves by displaying the curve for each element of each mineral.
- Use the button “Adjust (Manual)” to modify the calibration curve in case the calibration algorithm failed to set a proper calibration curveSelect the option “Apply to all masks”
- Press the button “Apply Standardization”
 to calibrate all maps and send the data to XMapTools
to calibrate all maps and send the data to XMapTools
- Check the calibrated maps and the merged map using the primary tree menu
- If the calibration quality is not good, select the category “Quanti” in the primary tree menu, then right-click on the name and select “Clear All” (optional). This will eliminate all the data in this category. You can do the same for the category “Merged”
Extracting Local bulk composition
- We need first to create a density map for the mask file used for calibration:
- Select a mask file in the secondary tree menu
- In the workspace “CALIBRATE”, press the button “Density”
 to automatically generate a density map
to automatically generate a density map - Enter average density values for each phase. Note that average values are automatically proposed if the mineral name used is available in the XMapTools database
- You can display the density map in that is listed under the category “Other” in the primary tree menu
- Display a merged map by selecting an oxide in the category “Merged” of the primary tree menu
- Select “Rectangle ROI”, then press the button “Add ROI”
 and immediately click and drag over the map to create the ROI
and immediately click and drag over the map to create the ROI - For this exercise, reshape the ROI to about ¾ of the map surface. If the editing mode is disabled, press once the “Ctrl” key and try again editing the shape of the ROI
- Save the local bulk composition
- Set the number of simulations to 100 and the shift value expressed in number of pixels to 20 and press the button “Calculate the uncertainties using Monte Carlo”

- Save the results and the figure (File > Save As…)
- Change the ROI to cover the entire map and export the new bulk composition. Compare the composition with the one previously obtained for the smaller domain
- Create a new merged map “garnet-excluded” containing only pixel compositions of biotite, plagioclase, quartz, sillimanite and sudoite and export the local bulk composition:
- Select the item “Biotite” in the category “Quanti” of the primary tree menu
- Press the button “Merge”

- Select all mineral except Garnet and press the button “Apply”
- Rename the merged map into “Merged_noGrt”
- Export a local bulk composition of the entire area
Structural formulas, thermometry & generating images
- Set the active workspace to “FUNCTIONS”
- In “Normalization & Structural Formula”, select “Function”, “Biotite” and “Bt (SF, 11-Ox. basis)”.
- Display the function description by clicking on the button

- Select “Biotite” in the category “Quanti” of the primary tree menu
- Press the button “Apply”
 to calculate the maps of structural formula for biotite
to calculate the maps of structural formula for biotite - Display the maps of structural formula “Biotite Bt (SF, 11-Ox. basis)” available in the category “Results” of the primary tree menu
- Generate an image showing the maps of Si, Ti and XMg in biotite:
- In the menu, select in the menu: Image > Multi-Selection Mode
- Select the maps Si, Ti_M2 and XMg. Press ctrl (Windows) or Command (Mac) to select several maps in the primary tree menu
- Once the maps are selected, select in the menu: Images > Add Multi-plot image
- Unfold the new image available under the category “Images” in the primary tree menu. Select each map and use the auto-contrast option
 or the live frequency polygon to adjust the lower and upper limits of the color bar. Note that the settings are saved when modifying an image
or the live frequency polygon to adjust the lower and upper limits of the color bar. Note that the settings are saved when modifying an image - To save the image, you can use the menu File > Save Image. Alternatively, you can copy the image into the clipboard and past it into your favorite vector graphics editor. In the menu, select: Edit > Copy Image
- Save your project; images are kept in the project file
- Calculate the maps of structural formula for garnet (with and without Fe3+) and plagioclase
- Calculate the maps of structural formula for garnet using a 8-cation basis
- Generate a new image containing the XMg maps of biotite and garnet:
- Activate the multi-selection mode (menu: Image > Multi-Selection Mode)
- In the menu “Image”, select the option “Add Multi-Layer Image (multi-scale)”
- Adjust the color scale and the color palette of each map in the image object
- Display the multi-layer image
- Select an other map (not an image) and change back the color palette to “XMap (CD)”. Note that this change does not affect the image
- Generate a new image containing maps of end-member fraction for almandine, pyrope, grossular and spessartine
- Generate an image of Mg in biotite and in garnet (in apfu):
- Select any result in “Biotite Bt (SF, 11-Ox. basis)”
- In the menu, select: Modules > Generator
- Type the following equation in the field that calculates Mg (apfu): “Mg = Mg_M1 + Mg_M2”
- Press the button “Generate”
 Activate the multi-selection mode
Activate the multi-selection mode - Select the maps Mg in garnet and Mg in biotite; both are expressed in apfu
- In the menu “Image”, select the option “Add Multi-Layer Image (shared-scale)”
- Use the live frequency polygon to adjust the lower and upper limits of the color bar
- Save the project
- Calculate temperature maps using Ti-in-biotite thermometry:
- In the workspace “FUNCTIONS”, activate the “Map mode” in the section “Thermobarometry and other methods”, select “Biotite” and “T.Bt (all calibrations)”
- Select “Biotite” in the category “Quanti”
- Press the button “Apply”
 available in the section “Thermobarometry and other methods”
available in the section “Thermobarometry and other methods” - Set the pressure to 6 kbar (0.6 GPa)
- Display the map T_H05
- Save a single image containing the temperature map T_H05
- Calculate temperature conditions using garnet-biotite thermometry:
- Select the merge map “Merged_RandomForest”
- Adjust the view using zoom
 and pan
and pan 
- In the workspace “FUNCTIONS”, activate the “Multi-equilibrium” mode in the section “Thermobarometry and other methods”, select “Biotite” or “Garnet” and “T.Bt (all calibrations)”
- Display the map FeOSelect “Circle ROI”, then press the button “Add ROI”

- Set the pressure to 6 kbar (0.6 GPa)
- Select a ROI for garnet and then a ROI for biotite
- Results are displayed in a table for all calibrations. Check the help function
 to obtain the references
to obtain the references - Save the results using the button
 save available above the table
save available above the table - Adjust the position of the ROI and extract the new temperature
- Click on the button “Reset ROI”

Sampling tools and chemical diagrams
- Export a transect chemical profile for the garnet end-member fractions:
- Display the map Xprp and adjust the lower and upper limits of the color bar
- In the menu, select Sampling > Transect and click on the image to define a transect from rim through core to rim
- Adjust the position of the transect (optional)
- Save the results for the selected end-member fraction. In the menu, select Sampling > Save Results > Single MapSelect Xsps, adjust the range of displayed values and save the results
- Select Xalm, adjust the range of displayed values and save the results
- Export an integrated profile (strip) for the fraction of pyrope in garnet
- Select the map Xprp and adjust the range of displayed values
- In the menu, select Sampling > Strip and click on the image to create rectangle
- Adjust the rotation and the position of the rectangle to sample from rim to core of a garnet grain
- Save the results and compare the profile obtained with the ones obtained using the transect tool
- Reset ROI using the button

- Generate a binary plot for biotite:
- Select any map of the structural formula of biotite
- In the menu select Modules > Data Vizualisation
- Display XMg vs TI_M2 and adjust the display by zooming on the figure
- Select a ROI using the button “Polygon”
available in “Explore”How many groups of biotite can be identified?
- Close the Data Vizualisation module to resume XMapTools
- Generate a ternary plot for plagioclase:
- Select any map of the structural formula of plagioclase
- In the menu select Modules > Data Vizualisation
- Select “Ternary” plot and the following variables: Xab for X , Xsan for Y and Xan for Z
- Use the “Polygon” tool
 to identify pixels locations and to find mixing pixels
to identify pixels locations and to find mixing pixels - Close the Data Vizualisation module to resume XMapTools
- Identify mixing pixels using intensity maps:
- Select any intensity map in the primary tree menu
- In the menu select Modules > Data Vizualisation
- Display a binary diagram Al vs Mg
- Use the “Polygon” tool
 to identify the minerals and the mixing pixels between them
to identify the minerals and the mixing pixels between them - Close the Data Vizualisation module to resume XMapTools
- You can also generate the same plot using quantitative data (e.g. merged map) to notice the differences with intensity maps. It should be kept in mind that intensity maps are uncorrected for ZAF effects!
Was this helpful?
17 / 0